https://bio.nikkeibp.co.jp/atcl/news/p1/23/04/25/10628/
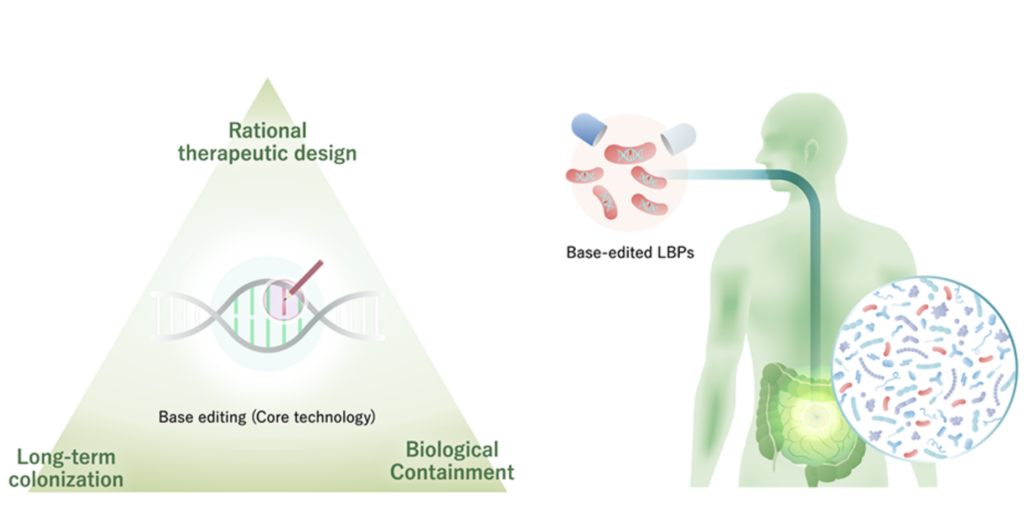
BioPalette is a Kobe University startup established in February 2017 based on research results from Kobe University. Professor Keiji Nishida and his research team at Kobe University’s Research Center for Advanced Biotechnology have developed “Target-AID” and “Target-G,” base-editing technologies that can rewrite bases without cutting DNA, among other genome editing technologies. Target-AID” uses deaminase as a catalytic enzyme to deaminate DNA and induces pinpoint point mutations. Target-G” uses glycosylase as a catalytic enzyme to deaminate DNA and induces random mutations in targeted regions. The unique feature of base-editing technology is that it can precisely edit DNA. In addition, it has the advantage of being less cytotoxic and easier to perform multiple edits compared to genome editing,” said Okumura.
In recent years, the advent of next-generation sequencers (NGS), which can analyze genomes inexpensively and quickly, and the establishment of experimental methods such as notobiotechnological analysis, in which specific bacteria are injected into the intestinal tracts of sterile animals, have stimulated research into the relationship between gut bacteria and disease.
BioPalette develops Living Biotherapeuic Products LBPs which are classified into three generations: “first generation,” in which wild-type bacteria are transplanted into patients; “second generation,” in which genetically modified intestinal bacteria (including foreign genes) are transplanted into patients; and “third generation,” in which bacteria modified by genome editing or base editing (not including foreign genes) are transplanted into patients. BioPalette is involved in the development of the third generation of LBPs. Specifically, wild-type E. coli and enterococci living in the intestinal tract are modified by base editing. The bacteria are then orally administered to replace the patient’s intestinal bacteria and produce a therapeutic effect. For example, stop codons could be introduced by base editing to inhibit the expression of target genes, or negative regulators could be introduced by base editing to increase the production of proteins. Currently, the company is considering LBPs for inflammatory bowel disease (IBD), cancer immunotherapy (using bacteria to improve therapeutic efficacy), central nervous system diseases, oral diseases, and skin diseases. The LBPs are based on E. coli and enterococci, which are endemic in the intestines and have a high association with diseases and for which evidence has been accumulated.